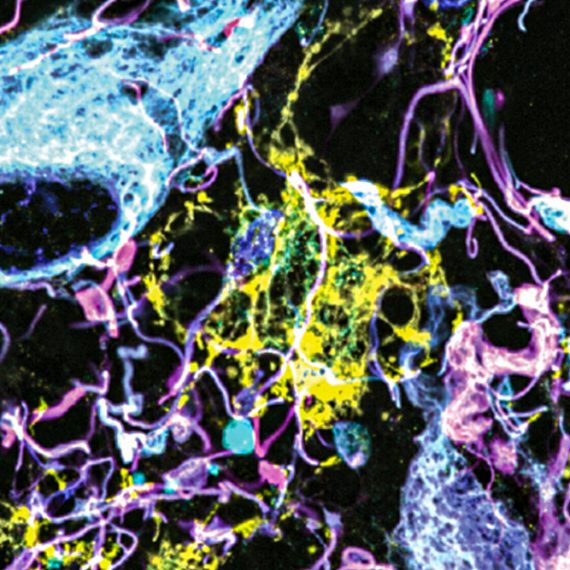
A Google map of the brain
New technologies allow researchers to map the brain’s component cells at a level of detail previously unimaginable.
At the start of the twentieth century, Santiago Ramón y Cajal’s drawings of brain cells under the microscope revealed a remarkable diversity of cell types within the brain. Through sketch after sketch, Cajal showed that the brain was not, as many believed, a web of self-similar material, but rather that it is composed of billions of cells of many different sizes, shapes, and interconnections.
Yet more than a hundred years later, we still do not know how many cell types make up the human brain. Despite decades of study, the challenge remains daunting, as the brain’s complexity has overwhelmed attempts to describe it systematically or to catalog its parts.
Now, however, this appears about to change, thanks to an explosion of new technical advances in areas ranging from DNA sequencing to microfluidics to computing and microscopy. For the first time, a parts list for the human brain appears to be within reach.
Why is this important? “Until we know all the cell types, we won’t fully understand how they are connected together,” explains McGovern Investigator Guoping Feng. “We know that the brain’s wiring is incredibly complicated, and that the connections are key to understanding how it works, but we don’t yet have the full picture. That’s what we are aiming for. It’s like making a Google map of the brain.”
Identifying the cell types is also important for understanding disease. As genetic risk factors for different disorders are identified, researchers need to know where they act within the brain, and which cell types and connections are disrupted as a result. “Once we know that, we can start to think about new therapeutic approaches,” says Feng, who is also an institute member of the Broad Institute, where he leads the neurobiology program at the Stanley Center for Psychiatric Disorders Research.
Drop by drop
In 2012, computational biologist Naomi Habib arrived from the Hebrew University of Jerusalem to join the labs of McGovern Investigator Feng Zhang and his collaborator Aviv Regev at the Broad Institute. Habib’s plan was to learn new RNA methods as they were emerging. “I wanted to use these powerful tools to understand this fascinating system that is our brain,” she says.
Her rationale was simple, at least in theory. All cells of an organism carry the same DNA instructions, but the instructions are read out differently in each cell type. Stretches of DNA corresponding to individual genes are copied, sometimes thousands of times, into RNA molecules that in turn direct the synthesis of proteins. Differences in which sequences get copied are what give cells their identities: brain cells express RNAs that encode brain proteins, while blood cells express different RNAs, and so on. A given cell can express thousands of genes, providing a molecular “fingerprint” for each cell type.
Analyzing these RNAs can provide a great deal of information about the brain, including potentially the identities of its constituent cell types. But doing this is not easy, because the different cell types are mixed together like salt and pepper within the brain. For many years, studying brain RNA meant grinding up the tissue—an approach that has been compared to studying smoothies to learn about fruit salad.
As methods improved, it became possible to study the tiny quantities of RNA contained within single cells. This opened the door to studying the difference between individual cells, but this required painstaking manipulation of many samples, a slow and laborious process.
A breakthrough came in 2015, with the development of automated methods based on microfluidics. One of these, known as dropseq (droplet-based sequencing), was pioneered by Steve McCarroll at Harvard, in collaboration with Regev’s lab at Broad. In this method, individual cells are captured in tiny water droplets suspended in oil. Vast numbers of droplets are automatically pumped through tiny channels, where each undergoes its own separate sequencing reactions. By running multiple samples in parallel, the machines can process tens of thousands of cells and billions of sequences, within hours rather than weeks or months. The power of the method became clear when in an experiment on mouse retina, the researchers were able to identify almost every cell type that had ever been described in the retina, effectively recapitulating decades of work in a single experiment.
Dropseq works well for many tissues, but Habib wanted to apply it to the adult brain, which posed a unique challenge. Mature neurons often bear elaborate branches that become intertwined like tree roots in a forest, making it impossible to separate individual cells without damage.
Nuclear option
So Habib turned to another idea. RNA is made in the nucleus before moving to the cytoplasm, and because nuclei are compact and robust it is easy to recover them intact in large numbers, even from difficult tissues such as brain. The amount of RNA contained in a single nucleus is tiny, and Habib didn’t know if it would be enough to be informative, but Zhang and Regev encouraged her to keep going. “You have to be optimistic,” she says. “You have to try.”
Fortunately, the experiment worked. In a paper with Zhang and Regev, she was able to isolate nuclei from newly formed neurons in the adult mouse hippocampus (a brain structure involved in memory), and by analyzing their RNA profiles individually she could order them in a series according to their age, revealing their developmental history from birth to maturity.
Now, after much further experimentation, Habib and her colleagues have managed to apply the droplet method to nuclei, making it possible for the first time to analyze huge numbers of cells from adult brain—at least ten times more than with previous methods.
This opens up many new avenues, including the study of human postmortem tissue, given that RNA in nuclei can survive for years in frozen samples. Habib is already starting to examine tissue taken at autopsy from patients with Alzheimer’s and other neurodegenerative diseases. “The neurons are degenerating, but the other cells around them could also be contributing to the degenerative process,” she says. “Now we have these tools, we can look at what happens during the progression of the disease.”
Computing cells
Once the sequencing is completed, the results are analyzed using sophisticated computational methods. When the results emerge, data from individual cells are visualized as colored dots, clustered on a graph according to their statistical similarities. But because the cells were dissociated at the start of the experiment, information about their appearance and origin within the brain is lost.
To find out how these abstract displays correspond to the visible cells of the brain, Habib teamed up with Yinqing Li, a former graduate student with Zhang who is now a postdoc in the lab of Guoping Feng. Li began with existing maps from the Allen Institute, a public repository with thousands of images showing expression patterns for individual genes within mouse brain. By comparing these maps with the molecular fingerprints from Habib’s nuclear RNA sequencing experiments, Li was able to make a map of where in the brain each cell was likely to have come from.
It was a good first step, but still not perfect. “What we really need,” he says, “is a method that allows us to see every RNA in individual cells. If we are studying a brain disease, we want to know which neurons are involved in the disease process, where they are, what they are connected to, and which special genes might be involved so that we can start thinking about how to design a drug that could alter the disease.”
Expanding horizons
So Li partnered with Asmamaw (Oz) Wassie, a graduate student in the lab of McGovern Investigator Ed Boyden, to tackle the problem. Wassie had previously studied bioengineering as an MIT undergraduate, where he had helped build an electronic “artificial nose” for detecting trace chemicals in air. With support from a prestigious Hertz Fellowship, he joined Boyden’s lab, where he is now working on the development of a method known as expansion microscopy.
In this method, a sample of tissue is embedded with a polymer that swells when water is added. The entire sample expands in all directions, allowing scientists to see fine details such as connections between neurons, using an ordinary microscope. Wassie recently helped develop a way to anchor RNA molecules to the polymer matrix, allowing them to be physically secured during the expansion process. Now, within the expanded samples he can see the individual molecules using a method called fluorescent in situ hybridization (FISH), in which each RNA appears as a glowing dot under the microscope. Currently, he can label only a handful of RNA types at once, but by using special sets of probes, applied sequentially, he thinks it will soon be possible to distinguish thousands of different RNA sequences.
“That will help us to see what each cell looks like, how they are connected to each other, and what RNAs they contain,” says Wassie. By combining this information with the RNA expression data generated by Li and Habib, it will be possible to reveal the organization and fine structure of complex brain areas and perhaps to identify new cell types that have not yet been recognized.
Looking ahead
Li plans to apply these methods to a brain structure known as the thalamic reticular nucleus (TRN) – a sheet of tissue, about ten neurons thick in mice, that sits on top of the thalamus and close to the cortex. The TRN is not well understood, but it is important for controlling sleep, attention and sensory processing, and it has caught the interest of Feng and other neuroscientists because it expresses a disproportionate number of genes implicated in disorders such as autism, attention deficit hyperactivity disorder, and intelligence deficits. Together with Joshua Levin’s group at Broad, Li has already used nuclear RNA sequencing to identify the cell types in the TRN, and he has begun to examine them within intact brain using the expansion techniques. “When you map these precise cell types back to the tissue, you can integrate the gene expression information with everything else, like electrophysiology, connectivity, morphology,” says Li. “Then we can start to ask what’s going wrong in disease.”
Meanwhile, Feng is already looking beyond the TRN, and planning how to scale the approach to other structures and eventually to the entire brain. He returns to the metaphor of a Google map. “Microscopic images are like satellite photos,” he says. “Now with expansion microscopy we can add another layer of information, like property boundaries and individual buildings. And knowing which RNAs are in each cell will be like seeing who lives in those buildings. I think this will completely change how we view the brain.”