Seeing more in expansion microscopy
A pair of new methods light up lipid membranes and let researchers see sets of proteins inside cells with high resolution.
In biology, seeing can lead to understanding, and researchers in Edward Boyden’s lab at MIT’s McGovern Institute are committed to bringing life into sharper focus. With a pair of new methods, they are expanding the capabilities of expansion microscopy—a high-resolution imaging technique the group introduced in 2015—so researchers everywhere can see more when they look at cells and tissues under a light microscope.

“We want to see everything, so we’re always trying to improve it,” says Boyden, the Y. Eva Tan Professor in Neurotechnology at MIT. “A snapshot of all life, down to its fundamental building blocks, is really the goal.” Boyden is also a Howard Hughes Medical Institute investigator and a member of the Yang Tan Collective at MIT.
With new ways of staining their samples and processing images, users of expansion microscopy can now see vivid outlines of the shapes of cells in their images and pinpoint the locations of many different proteins inside a single tissue sample with resolution that far exceeds that of conventional light microscopy. These advances, both reported in the journal Nature Communications, enable new ways of tracing the slender projections of neurons and visualizing spatial relationships between molecules that contribute to health and disease.
Expansion microscopy uses a water-absorbing hydrogel to physically expand biological tissues. After a tissue sample has been permeated by the hydrogel, it is hydrated. The hydrogel swells as it absorbs water, preserving the relative locations of molecules in the tissue as it gently pulls them away from one another. As a result, crowded cellular components appear separate and distinct when the expanded tissue is viewed under a light microscope. The approach, which can be performed using standard laboratory equipment, has made super-resolution imaging accessible to most research teams.
Since first developing expansion microscopy, Boyden and his team have continued to enhance the method—increasing its resolution, simplifying the procedure, devising new features, and integrating it with other tools.
Visualizing cell membranes
One of the team’s latest advances is a method called ultrastructural membrane expansion microscopy (umExM), which they described in the February 12 issue of Nature Communications. With it, biologists can use expansion microscopy to visualize the thin membranes that form the boundaries of cells and enclose the organelles inside them. These membranes, built mostly of molecules called lipids, have been notoriously difficult to densely label in intact tissues for imaging with light microscopy. Now, researchers can use umExM to study cellular ultrastructure and organization within tissues.
Tay Shin, a former graduate student in Boyden’s lab and a J. Douglas Tan Fellow in the Tan-Yang Center for Autism Research at MIT, led the development of umExM. “Our goal was very simple at first: Let’s label membranes in intact tissue, much like how an electron microscope uses osmium tetroxide to label membranes to visualize the membranes in tissue,” he says. “It turns out that it’s extremely hard to achieve this.”
The team first needed to design a label that would make the membranes in tissue samples visible under a light microscope. “We almost had to start from scratch,” Shin says. “We really had to think about the fundamental characteristics of the probe that is going to label the plasma membrane, and then think about how to incorporate them into expansion microscopy.” That meant engineering a molecule that would associate with the lipids that make up the membrane and link it to both the hydrogel used to expand the tissue sample and a fluorescent molecule for visibility.
After optimizing the expansion microscopy protocol for membrane visualization and extensively testing and improving potential probes, Shin found success one late night in the lab. He placed an expanded tissue sample on a microscope and saw sharp outlines of cells.
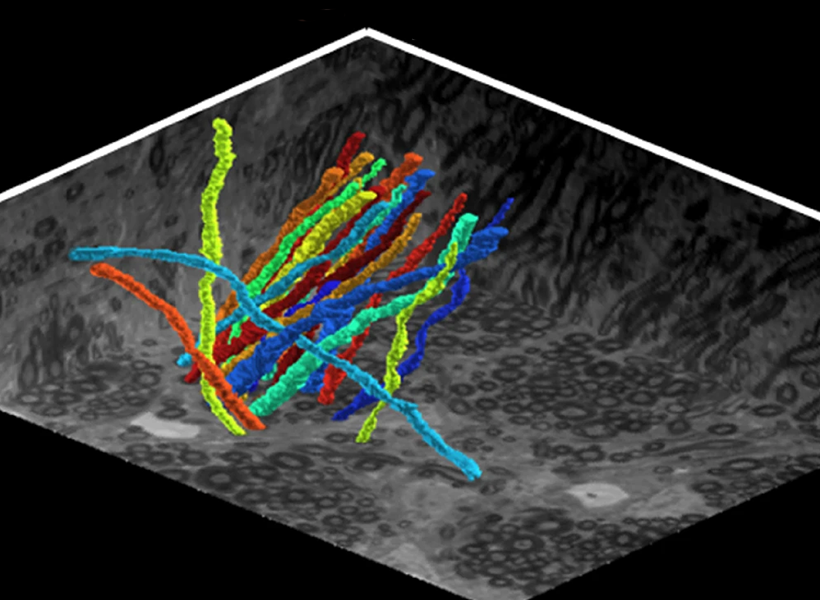
Because of the high resolution enabled by expansion, the method allowed Boyden’s team to identify even the tiny dendrites that protrude from neurons and clearly see the long extensions of their slender axons. That kind of clarity could help researchers follow individual neurons’ paths within the densely interconnected networks of the brain, the researchers say.
Boyden calls tracing these neural processes “a top priority of our time in brain science.” Such tracing has traditionally relied heavily on electron microscopy, which requires specialized skills and expensive equipment. Shin says that because expansion microscopy uses a standard light microscope, it is far more accessible to laboratories worldwide.
Shin and Boyden point out that users of expansion microscopy can learn even more about their samples when they pair the new ability to reveal lipid membranes with fluorescent labels that show where specific proteins are located. “That’s important, because proteins do a lot of the work of the cell, but you want to know where they are with respect to the cell’s structure,” Boyden says.
One sample, many proteins
To that end, researchers no longer have to choose just a few proteins to see when they use expansion microscopy. With a new method called multiplexed expansion revealing (multiExR), users can now label and see more than 20 different proteins in a single sample. Biologists can use the method to visualize sets of proteins, see how they are organized with respect to one another, and generate new hypotheses about how they might interact.
A key to the new method, reported November 9, 2024, in Nature Communications, is the ability to repeatedly link fluorescently labeled antibodies to specific proteins in an expanded tissue sample, image them, then strip these away and use a new set of antibodies to reveal a new set of proteins. Postdoctoral fellow Jinyoung Kang fine-tuned each step of this process, assuring tissue samples stayed intact and the labeled proteins produced bright signals in each round of imaging.
After capturing many images of a single sample, Boyden’s team faced another challenge: how to ensure those images were in perfect alignment so they could be overlaid with one another, producing a final picture that showed the precise positions of all of the proteins that had been labeled and visualized one by one.
Expansion microscopy lets biologists visualize some of cells’ tiniest features—but to find the same features over and over again during multiple rounds of imaging, Boyden’s team first needed to home in on a larger structure. “These fields of view are really tiny, and you’re trying to find this really tiny field of view in a gel that’s actually become quite large once you’ve expanded it,” explains Margaret Schroeder, a graduate student in Boyden’s lab who, with Kang, led the development of multiExR.
“Here’s one of the most famous receptors in all of neuroscience, hiding out in one of the most famous molecular hallmarks of pathology in neuroscience.” – Ed Boyden
To navigate to the right spot every time, the team decided to label the blood vessels that pass through each tissue sample and use these as a guide. To enable precise alignment, certain fine details also needed to consistently appear in every image; for this, the team labeled several structural proteins. With these reference points and customized imaging processing software, the team was able to integrate all of their images of a sample into one, revealing how proteins that had been visualized separately were arranged relative to one another.
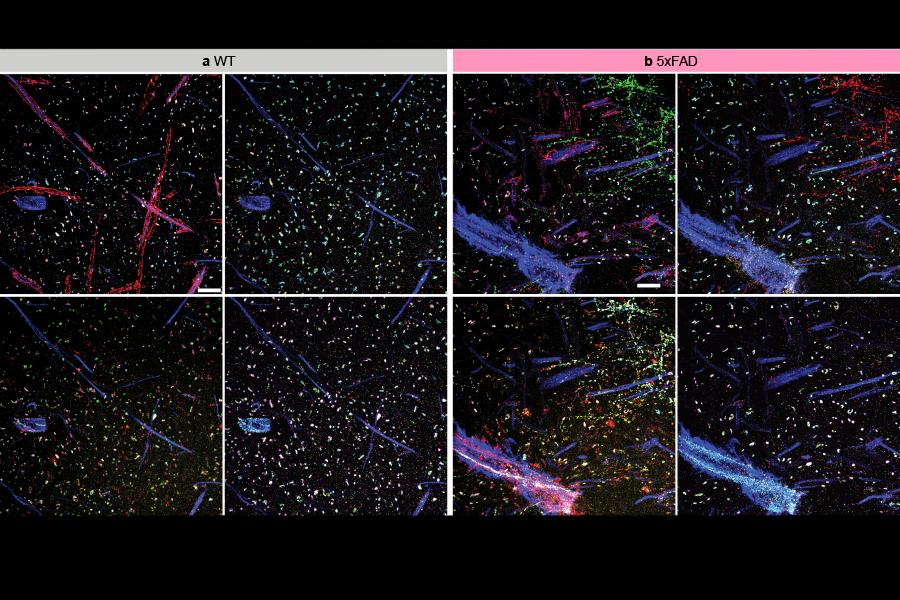
The team used multiExR to look at amyloid plaques—the aberrant protein clusters that notoriously develop in brains affected by Alzheimer’s disease. “We could look inside those amyloid plaques and ask, what’s inside of them? And because we can stain for many different proteins, we could do a high throughput exploration,” Boyden says. The team chose 23 different proteins to view in their images. The approach revealed some surprises, such as the presence of certain neurotransmitter receptors (AMPARs). “Here’s one of the most famous receptors in all of neuroscience, and there it is, hiding out in one of the most famous molecular hallmarks of pathology in neuroscience,” says Boyden. It’s unclear what role, if any, the receptors play in Alzheimer’s disease—but the finding illustrates how the ability to see more inside cells can expose unexpected aspects of biology and raise new questions for research.
Funding for this work came from MIT, Lisa Yang and Y. Eva Tan, John Doerr, the Open Philanthropy Project, the Howard Hughes Medical Institute, the US Army, Cancer Research UK, the New York Stem Cell Foundation, the National Institutes of Health, Lore McGovern, Good Ventures, Schmidt Futures. Samsung, MathWorks, the Collamore-Rogers Fellowship, the National Science Foundation, Alana Foundation USA, the Halis Family Foundation, Lester A. Gimpelson, Donald and Glenda Mattes, David B. Emmes, Thomas A. Stocky, Avni U. Shah, Kathleen Octavio, Good Ventures/Open Philanthropy, and the European Union’s Horizon 2020 program.