The new delivery platform works efficiently in cell models, and, with further development, could open up a new class of delivery methods for a wide range of molecular medicines — including those for gene editing and gene replacement. Existing delivery vehicles for these therapeutics can be inefficient and randomly integrate into the genome of cells, and some can stimulate unwanted immune reactions. SEND has the promise to overcome these limitations, which could open up new opportunities to deploy molecular medicine.
“The biomedical community has been developing powerful molecular therapeutics, but delivering them to cells in a precise and efficient way is challenging,” said CRISPR pioneer Feng Zhang, senior author on the study, core institute member at the Broad Institute, investigator at the McGovern Institute, and the James and Patricia Poitras Professor of Neuroscience at MIT. “SEND has the potential to overcome these challenges.” Zhang is also an investigator at the Howard Hughes Medical Institute and a professor in MIT’s Departments of Brain and Cognitive Sciences and Biological Engineering.
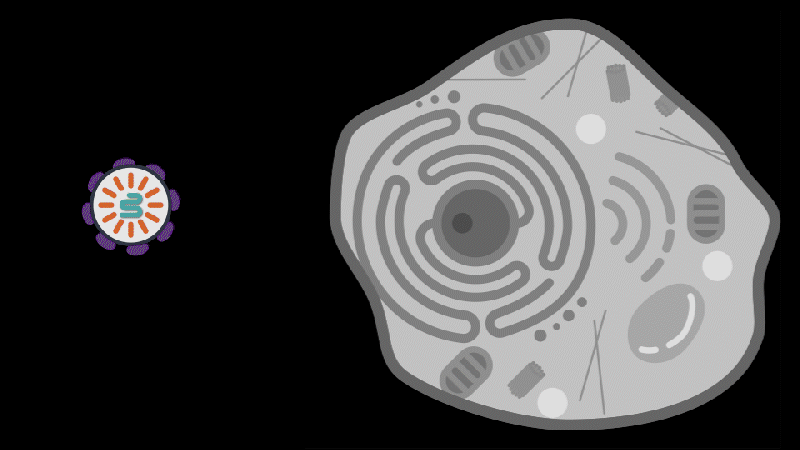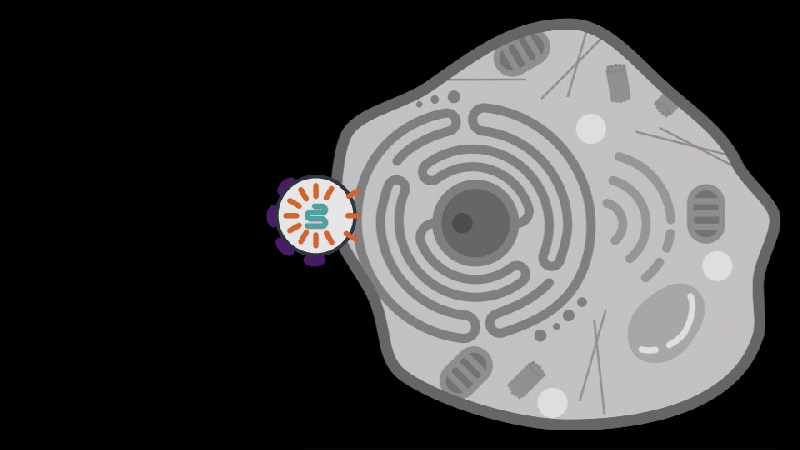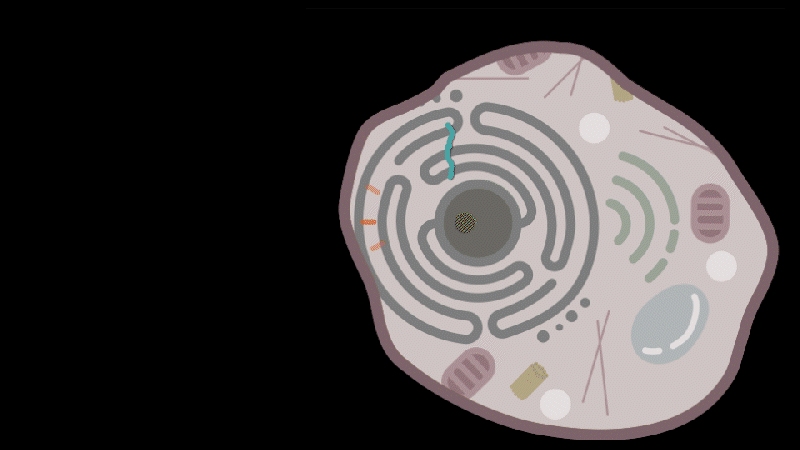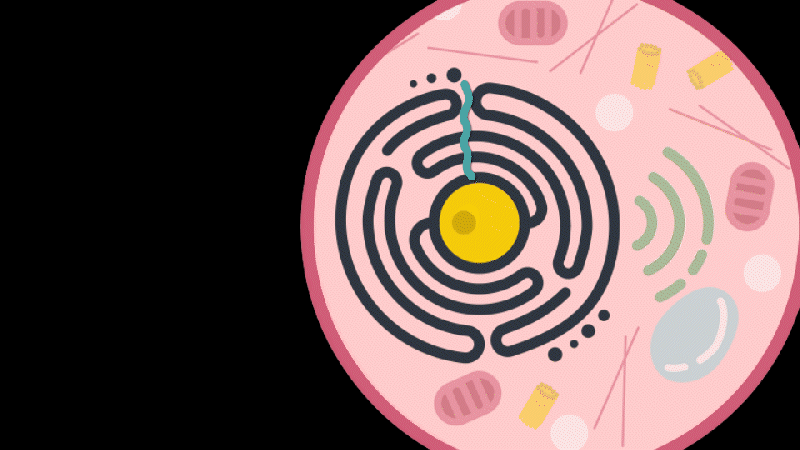
Reporting in Science, the team describes how SEND (Selective Endogenous eNcapsidation for cellular Delivery) takes advantage of molecules made by human cells. At the center of SEND is a protein called PEG10, which normally binds to its own mRNA and forms a spherical protective capsule around it. In their study, the team engineered PEG10 to selectively package and deliver other RNA. The scientists used SEND to deliver the CRISPR-Cas9 gene editing system to mouse and human cells to edit targeted genes.
First author Michael Segel, a postdoctoral researcher in Zhang’s lab, and Blake Lash, second author and a graduate student in the lab, said PEG10 is not unique in its ability to transfer RNA. “That’s what’s so exciting,” said Segel. “This study shows that there are probably other RNA transfer systems in the human body that can also be harnessed for therapeutic purposes. It also raises some really fascinating questions about what the natural roles of these proteins might be.”
Inspiration from within
The PEG10 protein exists naturally in humans and is derived from a “retrotransposon” — a virus-like genetic element — that integrated itself into the genome of human ancestors millions of years ago. Over time, PEG10 has been co-opted by the body to become part of the repertoire of proteins important for life.
Four years ago, researchers showed that another retrotransposon-derived protein, ARC, forms virus-like structures and is involved in transferring RNA between cells. Although these studies suggested that it might be possible to engineer retrotransposon proteins as a delivery platform, scientists had not successfully harnessed these proteins to package and deliver specific RNA cargoes in mammalian cells.
Knowing that some retrotransposon-derived proteins are able to bind and package molecular cargo, Zhang’s team turned to these proteins as possible delivery vehicles. They systematically searched through these proteins in the human genome for ones that could form protective capsules. In their initial analysis, the team found 48 human genes encoding proteins that might have that ability. Of these, 19 candidate proteins were present in both mice and humans. In the cell line the team studied, PEG10 stood out as an efficient shuttle; the cells released significantly more PEG10 particles than any other protein tested. The PEG10 particles also mostly contained their own mRNA, suggesting that PEG10 might be able to package specific RNA molecules.
Developing a modular system
To develop the SEND technology, the team identified the molecular sequences, or “signals,” in PEG10’s mRNA that PEG10 recognizes and uses to package its mRNA. The researchers then used these signals to engineer both PEG10 and other RNA cargo so that PEG10 could selectively package those RNAs. Next, the team decorated the PEG10 capsules with additional proteins, called “fusogens,” that are found on the surface of cells and help them fuse together.
By engineering the fusogens on the PEG10 capsules, researchers should be able to target the capsule to a particular kind of cell, tissue, or organ. As a first step towards this goal, the team used two different fusogens, including one found in the human body, to enable delivery of SEND cargo.
“By mixing and matching different components in the SEND system, we believe that it will provide a modular platform for developing therapeutics for different diseases,” said Zhang.
Advancing gene therapy
SEND is composed of proteins that are produced naturally in the body, which means it may not trigger an immune response. If this is demonstrated in further studies, the researchers say SEND could open up opportunities to deliver gene therapies repeatedly with minimal side effects. “The SEND technology will complement viral delivery vectors and lipid nanoparticles to further expand the toolbox of ways to deliver gene and editing therapies to cells,” said Lash.
Next, the team will test SEND in animals and further engineer the system to deliver cargo to a variety of tissues and cells. They will also continue to probe the natural diversity of these systems in the human body to identify other components that can be added to the SEND platform.
“We’re excited to keep pushing this approach forward,” said Zhang. “The realization that we can use PEG10, and most likely other proteins, to engineer a delivery pathway in the human body to package and deliver new RNA and other potential therapies is a really powerful concept.”
This work was made possible with support from the Simons Center for the Social Brain at MIT; National Institutes of Health Intramural Research Program; National Institutes of Health grants 1R01-HG009761 and 1DP1-HL141201; Howard Hughes Medical Institute; Open Philanthropy; G. Harold and Leila Y. Mathers Charitable Foundation; Edward Mallinckrodt, Jr. Foundation; Poitras Center for Psychiatric Disorders Research at MIT; Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT; Yang-Tan Center for Molecular Therapeutics at MIT; Lisa Yang; Phillips family; R. Metcalfe; and J. and P. Poitras.

