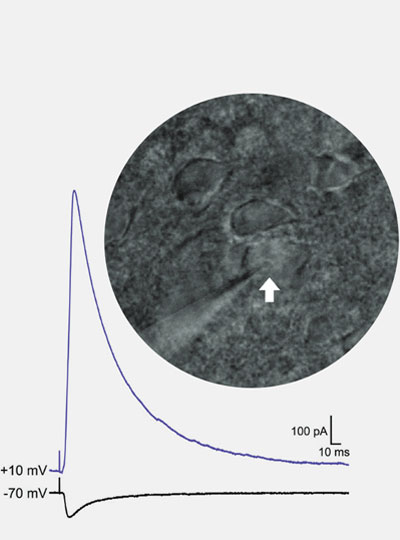
When we press our temples to soothe an aching head or rub an elbow after an unexpected blow, it often brings some relief. It is believed that pain-responsive cells in the brain quiet down when these neurons also receive touch inputs, say scientists at MIT’s McGovern Institute, who for the first time have watched this phenomenon play out in the brains of mice.
The team’s discovery, reported November 16, 2022, in the journal Science Advances, offers researchers a deeper understanding of the complicated relationship between pain and touch and could offer some insights into chronic pain in humans. “We’re interested in this because it’s a common human experience,” says McGovern Investigator Fan Wang. “When some part of your body hurts, you rub it, right? We know touch can alleviate pain in this way.” But, she says, the phenomenon has been very difficult for neuroscientists to study.
Modeling pain relief
Touch-mediated pain relief may begin in the spinal cord, where prior studies have found pain-responsive neurons whose signals are dampened in response to touch. But there have been hints that the brain was involved too. Wang says this aspect of the response has been largely unexplored, because it can be hard to monitor the brain’s response to painful stimuli amidst all the other neural activity happening there—particularly when an animal moves.
So while her team knew that mice respond to a potentially painful stimulus on the cheek by wiping their faces with their paws, they couldn’t follow the specific pain response in the animals’ brains to see if that rubbing helped settle it down. “If you look at the brain when an animal is rubbing the face, movement and touch signals completely overwhelm any possible pain signal,” Wang explains.
She and her colleagues have found a way around this obstacle. Instead of studying the effects of face-rubbing, they have focused their attention on a subtler form of touch: the gentle vibrations produced by the movement of the animals’ whiskers. Mice use their whiskers to explore, moving them back and forth in a rhythmic motion known as whisking to feel out their environment. This motion activates touch receptors in the face and sends information to the brain in the form of vibrotactile signals. The human brain receives the same kind of touch signals when a person shakes their hand as they pull it back from a painfully hot pan—another way we seek touch-mediate pain relief.
If you look at the brain when an animal is rubbing the face, movement and touch signals completely overwhelm any possible pain signal, says Wang.
Wang and her colleagues found that this whisker movement alters the way mice respond to bothersome heat or a poke on the face—both of which usually lead to face rubbing. “When the unpleasant stimuli were applied in the presence of their self-generated vibrotactile whisking…they respond much less,” she says. Sometimes, she says, whisking animals entirely ignore these painful stimuli.
In the brain’s somatosensory cortex, where touch and pain signals are processed, the team found signaling changes that seem to underlie this effect. “The cells that preferentially respond to heat and poking are less frequently activated when the mice are whisking,” Wang says. “They’re less likely to show responses to painful stimuli.” Even when whisking animals did rub their faces in response to painful stimuli, the team found that neurons in the brain took more time to adopt the firing patterns associated with that rubbing movement. “When there is a pain stimulation, usually the trajectory the population dynamics quickly moved to wiping. But if you already have whisking, that takes much longer,” Wang says.
Wang notes that even in the fraction of a second before provoked mice begin rubbing their faces, when the animals are relatively still, it can be difficult to sort out which brain signals are related to perceiving heat and poking and which are involved in whisker movement. Her team developed computational tools to disentangle these, and are hoping other neuroscientists will use the new algorithms to make sense of their own data.
Whisking’s effects on pain signaling seem to depend on dedicated touch-processing circuitry that sends tactile information to the somatosensory cortex from a brain region called the ventral posterior thalamus. When the researchers blocked that pathway, whisking no longer dampened the animals’ response to painful stimuli. Now, Wang says, she and her team are eager to learn how this circuitry works with other parts of the brain to modulate the perception and response to painful stimuli.
Wang says the new findings might shed light on a condition called thalamic pain syndrome, a chronic pain disorder that can develop in patients after a stroke that affects the brain’s thalamus. “Such strokes may impair the functions of thalamic circuits that normally relay pure touch signals and dampen painful signals to the cortex,” she says.