Look around, and you’ll see it everywhere: the way trees form branches, the way cities divide into neighborhoods, the way the brain organizes into regions. Nature loves modularity—a limited number of self-contained units that combine in different ways to perform many functions. But how does this organization arise? Does it follow a detailed genetic blueprint, or can these structures emerge on their own?
A new study from McGovern Associate Investigator Ila Fiete suggests a surprising answer.
In findings published today in Nature, Fiete, a professor of brain and cognitive sciences and director of the K. Lisa Yang Integrative Computational Neuroscience (ICoN) Center at MIT, reports that a mathematical model called peak selection can explain how modules emerge without strict genetic instructions. Her team’s findings, which apply to brain systems and ecosystems, help explain how modularity occurs across nature, no matter the scale.
Joining two big ideas
“Scientists have debated how modular structures form. One hypothesis suggests that various genes are turned on at different locations to begin or end a structure. This explains how insect embryos develop body segments, with genes turning on or off at specific concentrations of a smooth chemical gradient in the insect egg,” says Fiete, who is the senior author of the paper. Mikail Khona, a former graduate student and K. Lisa Yang ICoN Center Graduate Fellow, and postdoctoral associate Sarthak Chandra also led the study.
Another idea, inspired by mathematician Alan Turing, suggests that a structure could emerge from competition—small-scale interactions can create repeating patterns, like the spots on a cheetah or the ripples in sand dunes.
Both ideas work well in some cases, but fail in others. The new research suggests that nature need not pick one approach over the other. The authors propose a simple mathematical principle called peak selection, showing that when a smooth gradient is paired with local interactions that are competitive, modular structures emerge naturally. “In this way, biological systems can organize themselves into sharp modules without detailed top-down instruction,” says Chandra.
Modular systems in the brain
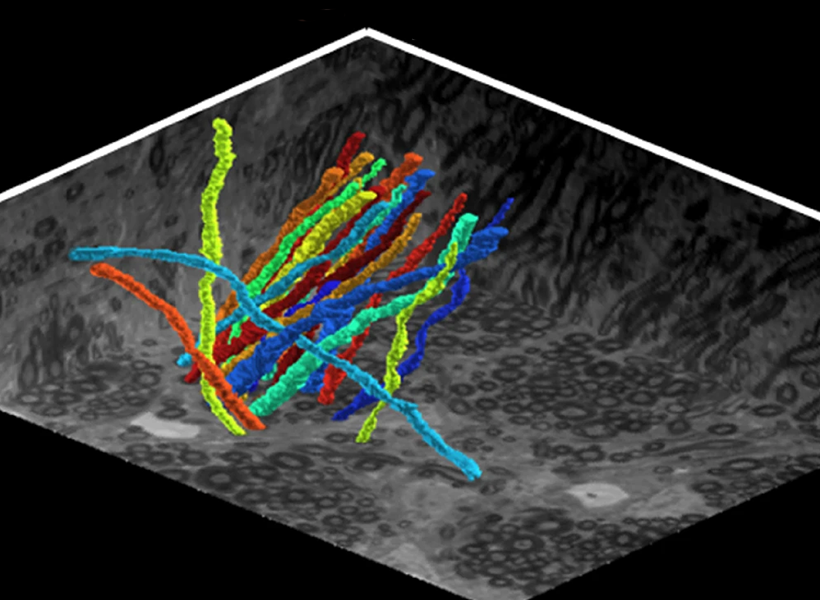
The researchers tested their idea on grid cells, which play a critical role in spatial navigation as well as the storage of episodic memories. Grid cells fire in a repeating triangular pattern as animals move through space, but they don’t all work at the same scale—they are organized into distinct modules, each responsible for mapping space at slightly different resolutions.
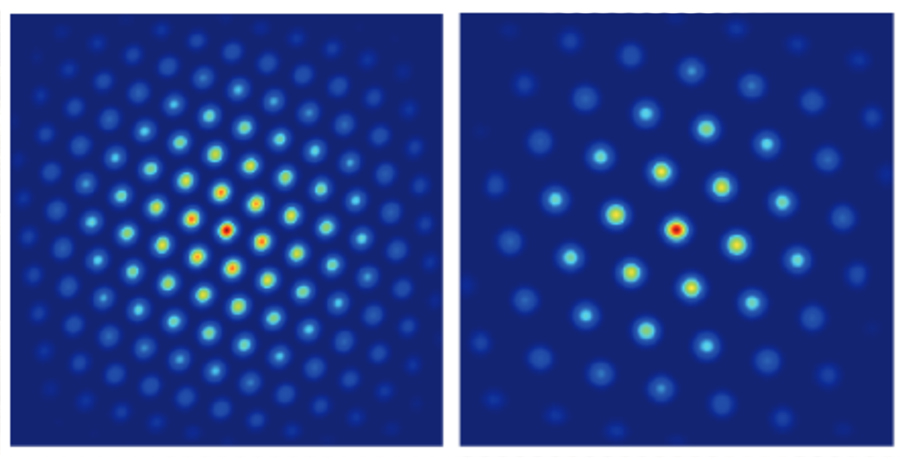
No one knows how these modules form, but Fiete’s model shows that gradual variations in cellular properties along one dimension in the brain, combined with local neural interactions, could explain the entire structure. The grid cells naturally sort themselves into distinct groups with clear boundaries, without external maps or genetic programs telling them where to go. “Our work explains how grid cell modules could emerge. The explanation tips the balance toward the possibility of self-organization. It predicts that there might be no gene or intrinsic cell property that jumps when the grid cell scale jumps to another module,” notes Khona.
Modular systems in nature
The same principle applies beyond neuroscience. Imagine a landscape where temperatures and rainfall vary gradually over a space. You might expect species to be spread and also vary smoothly over this region. But in reality, ecosystems often form species clusters with sharp boundaries—distinct ecological “neighborhoods” that don’t overlap.
Fiete’s study suggests why: Local competition, cooperation, and predation between species interact with the global environmental gradients to create natural separations, even when the underlying conditions change gradually. This phenomenon can be explained using peak selection—and suggests that the same principle that shapes brain circuits could also be at play in forests and oceans.
A self-organizing world
One of the researchers’ most striking findings is that modularity in these systems is remarkably robust. Change the size of the system, and the number of modules stays the same, they just scale up or down. That means a mouse brain and a human brain could use the same fundamental rules to form their navigation circuits, just at different sizes.
The model also makes testable predictions. If it’s correct, grid cell modules should follow simple spacing ratios. In ecosystems, species distributions should form distinct clusters even without sharp environmental shifts.
Fiete notes that their work adds another conceptual framework to biology. “Peak selection can inform future experiments, not only in grid cell research but across developmental biology.”