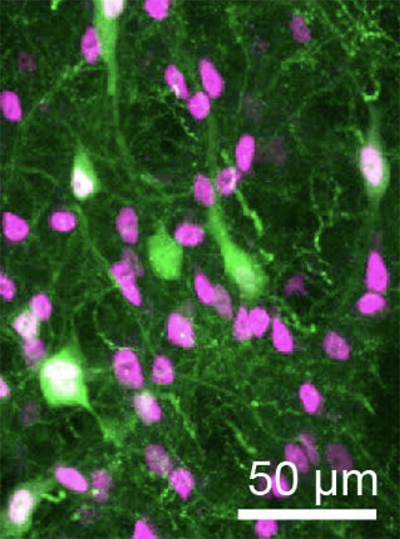
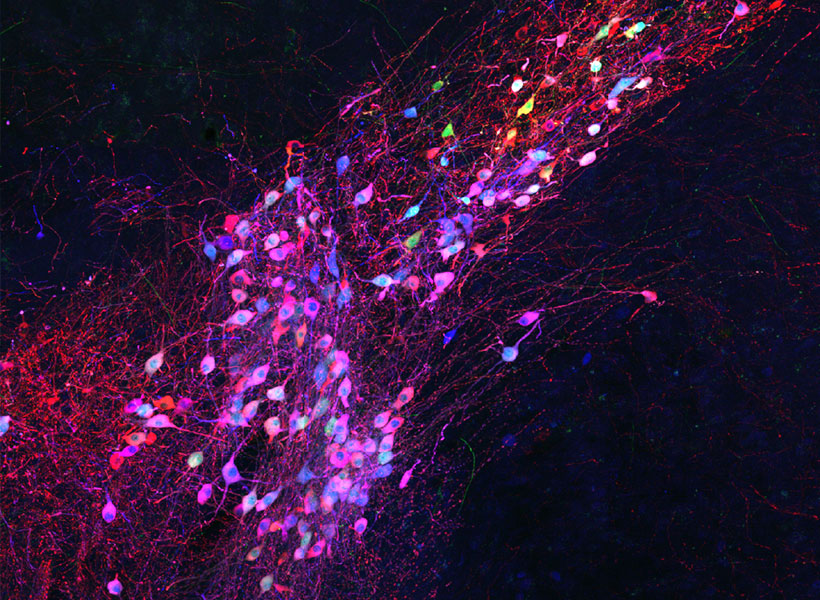
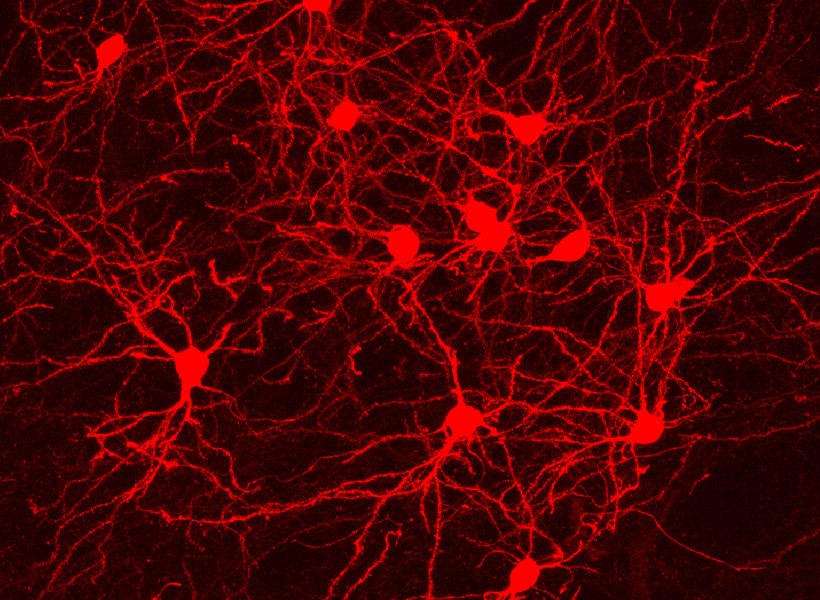
Mark Harnett, an associate professor at MIT, still remembers the first time he saw electrical activity spiking from a living neuron.
He was a senior at Reed College and had spent weeks building a patch clamp rig — an experimental setup with an electrode that can be used to gently probe a neuron and measure its electrical activity.
“The first time I stuck one of these electrodes onto one of these cells and could see the electrical activity happening in real time on the oscilloscope, I thought, ‘Oh my God, this is what I’m going to do for the rest of my life. This is the coolest thing I’ve ever seen!’” Harnett says.
Harnett, who recently earned tenure in MIT’s Department of Brain and Cognitive Sciences, now studies the electrical properties of neurons and how these properties enable neural circuits to perform the computations that give rise to brain functions such as learning, memory, and sensory perception.
“My lab’s ultimate goal is to understand how the cortex works,” Harnett says. “What are the computations? How do the cells and the circuits and the synapses support those computations? What are the molecular and structural substrates of learning and memory? How do those things interact with circuit dynamics to produce flexible, context-dependent computation?”
“We go after that by looking at molecules, like synaptic receptors and ion channels, all the way up to animal behavior, and building theoretical models of neural circuits,” he adds.
Influence on the mind
Harnett’s interest in science was sparked in middle school, when he had a teacher who made the subject come to life. “It was middle school science, which was a lot of just mixing random things together. It wasn’t anything particularly advanced, but it was really fun,” he says. “Our teacher was just super encouraging and inspirational, and she really sparked what became my lifelong interest in science.”
When Harnett was 11, his father got a new job at a technology company in Minneapolis and the family moved from New Jersey to Minnesota, which proved to be a difficult adjustment. When choosing a college, Harnett decided to go far away, and ended up choosing Reed College, a school in Portland, Oregon, that encourages a great deal of independence in both academics and personal development.
“Reed was really free,” he recalls. “It let you grow into who you wanted to be, and try things, both for what you wanted to do academically or artistically, but also the kind of person you wanted to be.”
While in college, Harnett enjoyed both biology and English, especially Shakespeare. His English professors encouraged him to go into science, believing that the field needed scientists who could write and think creatively. He was interested in neuroscience, but Reed didn’t have a neuroscience department, so he took the closest subject he could find — a course in neuropharmacology.
“That class totally blew my mind. It was just fascinating to think about all these pharmacological agents, be they from plants or synthetic or whatever, influencing how your mind worked,” Harnett says. “That class really changed my whole way of thinking about what I wanted to do, and that’s when I decided I wanted to become a neuroscientist.”
For his senior research thesis, Harnett joined an electrophysiology lab at Oregon Health Sciences University (OHSU), working with Professor Larry Trussell, who studies synaptic transmission in the auditory system. That lab was where he first built and used a patch clamp rig to measure neuron activity.
After graduating from college, he spent a year as a research technician in a lab at the University of Minnesota, then returned to OHSU to work in a different research lab studying ion channels and synaptic physiology. Eventually he decided to go to graduate school, ending up at the University of Texas at Austin, where his future wife was studying public policy.
For his PhD research, he studied the neurons that release the neuromodulator dopamine and how they are affected by drugs of abuse and addiction. However, once he finished his degree, he decided to return to studying the biophysics of computation, which he pursued during a postdoc at the Howard Hughes Medical Institute Janelia Research Campus with Jeff Magee.
A broad approach
When he started his lab at MIT’s McGovern Institute in 2015, Harnett set out to expand his focus. While the physiology of ion channels and synapses forms the basis of much of his lab’s work, they connect these processes to neuronal computation, cortical circuit operation, and higher-level cognitive functions.
Electrical impulses that flow between neurons, allowing them to communicate with each other, are produced by ion channels that control the flow of ions such as potassium and sodium. In a 2021 study, Harnett and his students discovered that human neurons have a much smaller number of these channels than expected, compared to the neurons of other mammals.
This reduction in density may have evolved to help the brain operate more efficiently, allowing it to divert resources to other energy-intensive processes that are required to perform complex cognitive tasks. Harnett’s lab has also found that in human neurons, electrical signals weaken as they flow along dendrites, meaning that small sections of dendrites can form units that perform individual computations within a neuron.
Harnett’s lab also recently discovered, to their surprise, that the adult brain contains millions of “silent synapses” — immature connections that remain inactive until they’re recruited to help form new memories. The existence of these synapses offers a clue to how the adult brain is able to continually form new memories and learn new things without having to modify mature synapses.
Many of these projects fall into areas that Harnett didn’t necessarily envision himself working on when he began his faculty career, but they naturally grew out of the broad approach he wanted to take to studying the cortex. To that end, he sought to bring people to the lab who wanted to work at different levels — from molecular physiology up to behavior and computational modeling.
As a postdoc studying electrophysiology, Harnett spent most of his time working alone with his patch clamp device and two-photon microscope. While that type of work still goes on his lab, the overall atmosphere is much more collaborative and convivial, and as a mentor, he likes to give his students broad leeway to come up with their own projects that fit in with the lab’s overall mission.
“I have this incredible, dynamic group that has been really great to work with. We take a broad approach to studying the cortex, and I think that’s what makes it fun,” he says. “Working with the folks that I’ve been able to recruit — grad students, techs, undergrads, and postdocs — is probably the thing that really matters the most to me.”