Language is a uniquely human ability that allows us to build vibrant pictures of non-existent places (think Wonderland or Westeros). How does the brain build mental worlds from words? Can machines do the same? Can we recover this ability after brain injury? These questions require an understanding of how the brain processes language, a fascination for Ev Fedorenko.
“I’ve always been interested in language. Early on, I wanted to found a company that teaches kids languages that share structure — Spanish, French, Italian — in one go,” says Fedorenko, an associate investigator at the McGovern Institute and an assistant professor in brain and cognitive sciences at MIT.
Her road to understanding how thoughts, ideas, emotions, and meaning can be delivered through sound and words became clear when she realized that language was accessible through cognitive neuroscience.
Early on, Fedorenko made a seminal finding that undermined dominant theories of the time. Scientists believed a single network was extracting meaning from all we experience: language, music, math, etc. Evolving separate networks for these functions seemed unlikely, as these capabilities arose recently in human evolution.
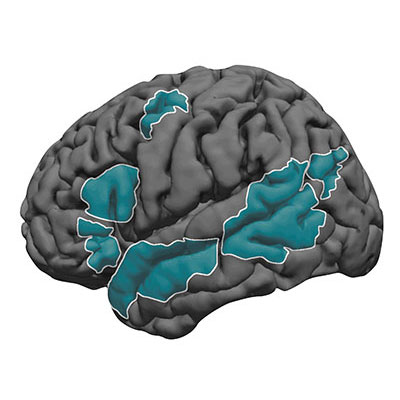
But when Fedorenko examined brain activity in subjects while they read or heard sentences in the MRI, she found a network of brain regions that is indeed specialized for language.
“A lot of brain areas, like motor and social systems, were already in place when language emerged during human evolution,” explains Fedorenko. “In some sense, the brain seemed fully occupied. But rather than co-opt these existing systems, the evolution of language in humans involved language carving out specific brain regions.”
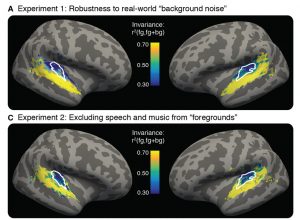
Different aspects of language recruit brain regions across the left hemisphere, including Broca’s area and portions of the temporal lobe. Many believe that certain regions are involved in processing word meaning while others unpack the rules of language. Fedorenko and colleagues have however shown that the entire language network is selectively engaged in linguistic tasks, processing both the rules (syntax) and meaning (semantics) of language in the same brain areas.
Semantic Argument
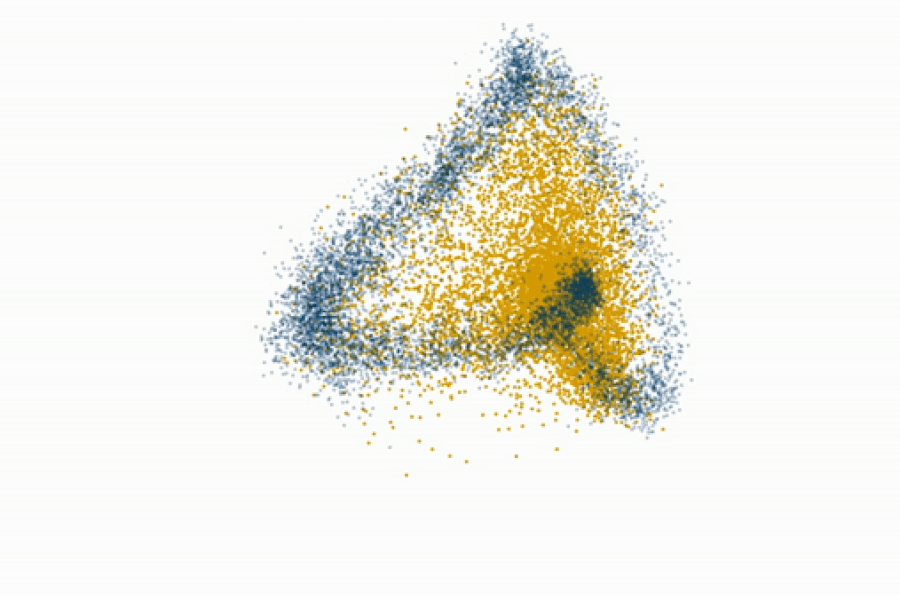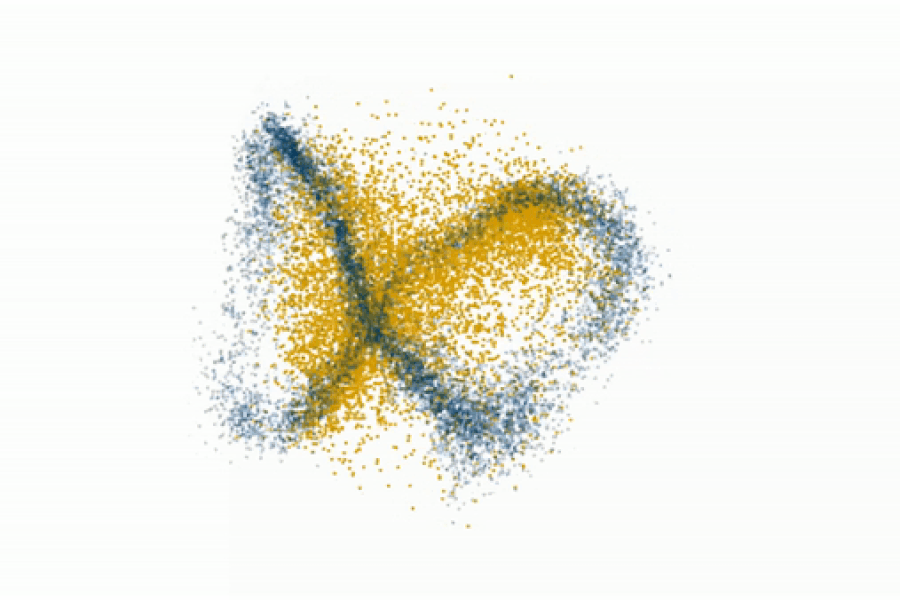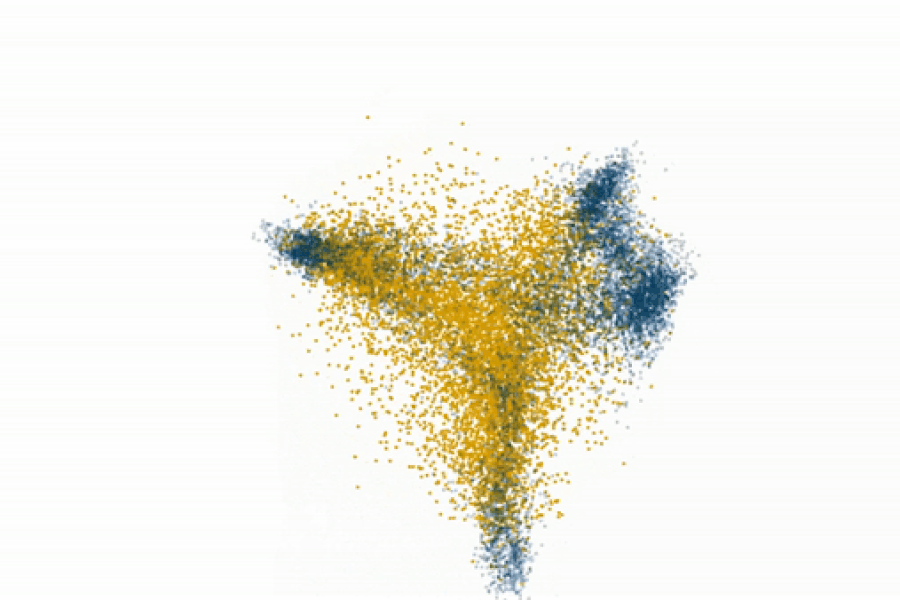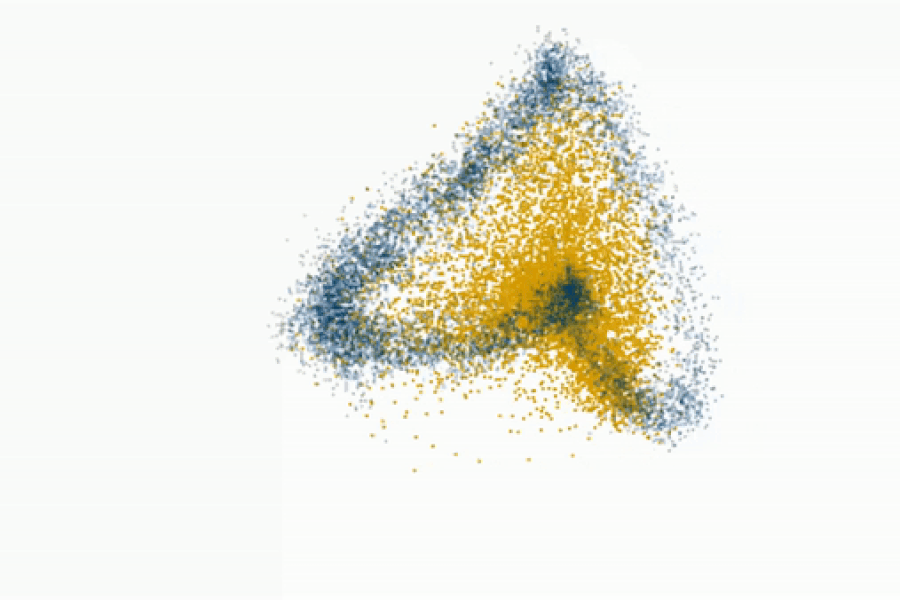
Fedorenko’s lab even challenges the prevailing view that syntax is core to language processing. By gradually degrading sentence structure through local word swaps (see figure), they found that language regions still respond strongly to these degraded sentences, deciphering meaning from them, even as syntax, or combinatorial rules, disappear.

“A lot of focus in language research has been on structure-building, or building a type of hierarchical graph of the words in a sentence. But actually the language system seems optimized and driven to find rich, representational meaning in a string of words processed together,” explains Fedorenko.
Computing Language
When asked about emerging areas of research, Fedorenko points to the data structures and algorithms underlying linguistic processing. Modern computational models can perform sophisticated tasks, including translation, ever more effectively. Consider Google translate. A decade ago, the system translated one word at a time with laughable results. Now, instead of treating words as providing context for each other, the latest artificial translation systems are performing more accurately. Understanding how they resolve meaning could be very revealing.
“Maybe we can link these models to human neural data to both get insights about linguistic computations in the human brain, and maybe help improve artificial systems by making them more human-like,” says Fedorenko.
She is also trying to understand how the system breaks down, how it over-performs, and even more philosophical questions. Can a person who loses language abilities (with aphasia, for example) recover — a very relevant question given the language-processing network occupies such specific brain regions. How are some unique people able to understand 10, 15 or even more languages? Do we need words to have thoughts?
Using a battery of approaches, Fedorenko seems poised to answer some of these questions.