Four MIT scientists are among the 20 recipients of the 2021 Academy Honors for major contributions to science, the National Academy of Sciences (NAS) announced at its annual meeting. The individuals are recognized for their “extraordinary scientific achievements in a wide range of fields spanning the physical, biological, social, and medical sciences.”
The awards recognize: Pablo Jarillo-Herrero, for contributions to the fields of nanoscience and nanotechnology through his discovery of correlated insulator behavior and unconventional superconductivity in magic-angle graphene superlattices; Aviv Regev, for using interdisciplinary information or techniques to solve a contemporary challenge; Susan Solomon, for contributions to understanding and communicating the causes of ozone depletion and climate change; and Feng Zhang, for pioneering achievements developing CRISPR tools with the potential to diagnose and treat disease.
Pablo Jarillo-Herrero: Award for Scientific Discovery
Pablo Jarillo-Herrero, a Cecil and Ida Green Professor of Physics, is the recipient of the NAS Award for Scientific Discovery for his pioneering developments in nanoscience and nanotechnology, which is presented to scientists in the fields of astronomy, materials science, or physics. His findings expand nanoscience by demonstrating for the first time that orientation can be used to dramatically control nanomaterial properties and to design new nanomaterials. This work lays the groundwork for developing a whole new family of 2D materials and has had a transformative impact on the field and on condensed-matter physics.
The biannual award recognizes “an accomplishment or discovery in basic research, achieved within the previous five years, that is expected to have a significant impact on one or more of the following fields: astronomy, biochemistry, biophysics, chemistry, materials science, or physics.”
In 2018, his research group discovered that by rotating two layers of graphene relative to each other by a magic angle, the bilayer material can be turned from a metal into an electrical insulator or even a superconductor. This discovery has fostered new theoretical and experimental research, inspiring the interest of technologists in nanoelectronics. The result is a new field in condensed-matter physics that has the potential to result in materials that conduct electricity without resistance at room temperature.
Aviv Regev: James Prize in Science and Technology Integration
Aviv Regev, who is a professor of biology, a core member of the Broad Institute of Harvard and MIT, a member of the Koch Institute, and a Howard Hughes Medical Institute investigator has been selected for the inaugural James Prize in Science and Technology Integration, along with Harvard Medical School Professor Allon Kelin, for “their concurrent development of now widely adopted massively parallel single-cell genomics to interrogate the gene expression profiles that define, at the level of individual cells, the distinct cell types in metazoan tissues, their developmental trajectories, and disease states, which integrated tools from molecular biology, engineering, statistics, and computer science.”
The prize recognizes individuals “who are able to adopt or adapt information or techniques from outside their fields” to “solve a major contemporary challenge not addressable from a single disciplinary perspective.”
Regev is credited with forging new ways to unite the disciplines of biology, computational science, and engineering as a pioneer in the field of single-cell biology, including developing some of its core experimental and analysis tools, and their application to discover cell types, states, programs, environmental responses, development, tissue locations, and regulatory circuits, and deploying these to assemble cellular atlases of the human body that illuminate mechanisms of disease with remarkable fidelity.
Susan Solomon: Award for Chemistry in Service to Society
Susan Solomon, the Lee and Geraldine Martin Professor of Environmental Studies in the Department of Earth, Atmospheric and Planetary Sciences who holds a secondary appointment in the Department of Chemistry, is the recipient of the Award for Chemistry in Service to Society for “influential and incisive application of atmospheric chemistry to understand our most critical environmental issues — ozone layer depletion and climate change — and for her effective communication of environmental science to leaders to facilitate policy changes.”
The award is given biannually for “contributions to chemistry, either in fundamental science or its application, that clearly satisfy a societal need.”
Solomon is globally recognized as a leader in atmospheric science, notably for her insights in explaining the cause of the Antarctic ozone “hole.” She and her colleagues have made important contributions to understanding chemistry-climate coupling, including pioneering research on the irreversibility of global warming linked to anthropogenic carbon dioxide emissions, and on the influence of the ozone hole on the climate of the southern hemisphere.
Her work has had an enormous effect on policy and society, including the transition away from ozone-depleting substances and to environmentally benign chemicals. The work set the stage for the Paris Agreement on climate, and she continues to educate policymakers, the public, and the next generation of scientists.
Feng Zhang: Richard Lounsbery Award
Feng Zhang, who is the James and Patricia Poitras Professor of Neuroscience at MIT, an investigator at the McGovern Institute for Brain Research and the Howard Hughes Medical Institute, a professor of brain and cognitive sciences and biological engineering at MIT, and a core member of the Broad Institute of MIT and Harvard, is the recipient of the Richard Lounsbery Award for pioneering CRISPR-mediated genome editing.
The award recognizes “extraordinary scientific achievement in biology and medicine” as well as stimulating research and encouraging reciprocal scientific exchanges between the United States and France.
Zhang continues to lead the field through the discovery of novel CRISPR systems and their development as molecular tools with the potential to diagnose and treat disease, such as disorders affecting the nervous system. His contributions in genome engineering, as well as his earlier work developing optogenetics, are enabling a deeper understanding of behavioral neural circuits and advances in gene therapy for treating disease.
In addition, Zhang has championed the open sharing of the technologies he has developed through extensive resource sharing. The tools from his lab are being used by thousands of scientists around the world to accelerate research in nearly every field of the life sciences. Even as biomedical researchers around the world adopt Zhang’s discoveries and his tools enter the clinic to treat genetic diseases, he continues to innovate and develop new technologies to advance science.
The National Academy of Sciences is a private, nonprofit society of distinguished scholars, established in 1863 by the U.S. Congress. The NAS is charged with providing independent, objective advice to the nation on matters related to science and technology as well as encouraging education and research, recognize outstanding contributions to knowledge, and increasing public understanding in matters of science, engineering, and medicine. Winners received their awards, which include a monetary prize, during a virtual ceremony at the 158th NAS Annual Meeting.
This story is a modified compilation from several National Academy of Sciences press releases.


 View the interactive version of this story in our
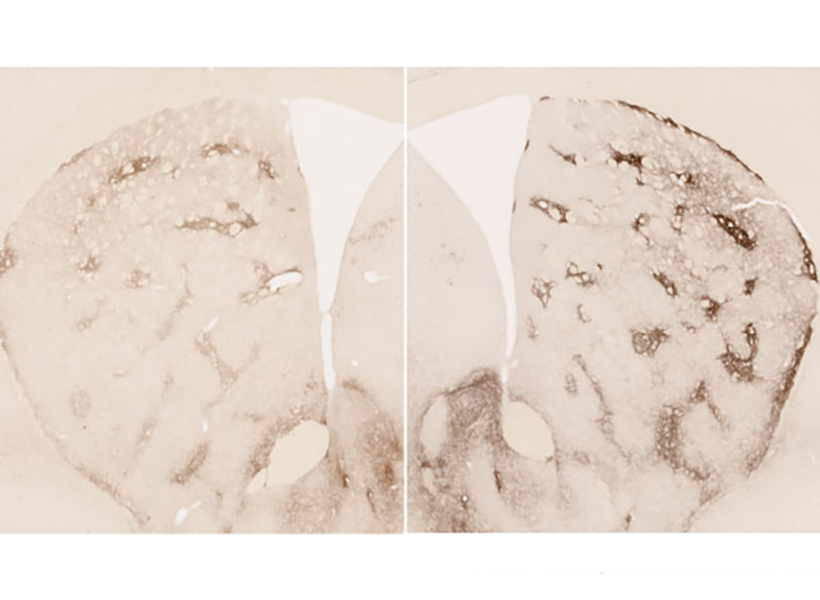
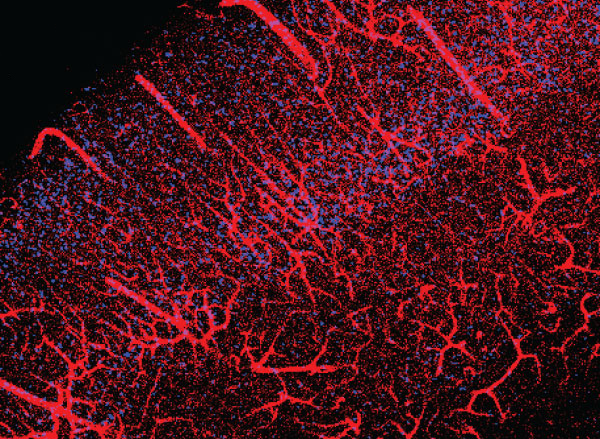
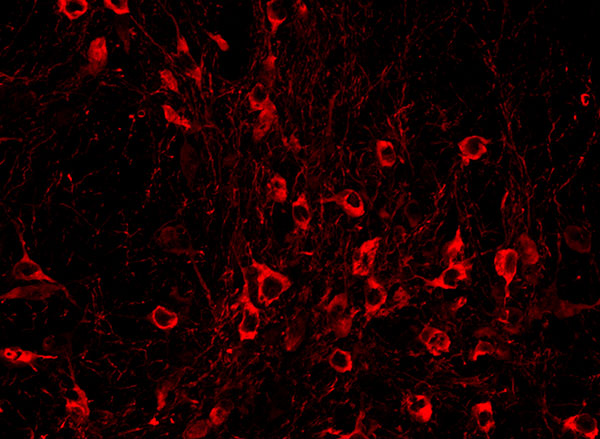
View the interactive version of this story in our  Taking advantage of its pernicious spread, neuroscientists use a modified version of the rabies virus to introduce a fluorescent protein to infected cells and visualize their connections (above). As a graduate student in Edward Callaway’s lab at the Salk Institute for Biological Studies, Wickersham figured out how to limit the virus’s passage through the nervous system, allowing it to access cells that are directly connected to the neuron it initially infects, but go no further. Rabies virus travels across synapses in the opposite direction of neuronal signals, so researchers can deliver it to a single cell or set of cells, then see exactly where those cells’ inputs are coming from.
Taking advantage of its pernicious spread, neuroscientists use a modified version of the rabies virus to introduce a fluorescent protein to infected cells and visualize their connections (above). As a graduate student in Edward Callaway’s lab at the Salk Institute for Biological Studies, Wickersham figured out how to limit the virus’s passage through the nervous system, allowing it to access cells that are directly connected to the neuron it initially infects, but go no further. Rabies virus travels across synapses in the opposite direction of neuronal signals, so researchers can deliver it to a single cell or set of cells, then see exactly where those cells’ inputs are coming from. In Feng’s lab, research scientist Martin Wienisch is working to make it easier to control this aspect of delivery. Rather than relying on the genetic makeup of an entire animal to determine where a virally-transported gene is switched on, instructions can be programmed directly into the virus, borrowing regulatory sequences that cells already know how to interpret.
In Feng’s lab, research scientist Martin Wienisch is working to make it easier to control this aspect of delivery. Rather than relying on the genetic makeup of an entire animal to determine where a virally-transported gene is switched on, instructions can be programmed directly into the virus, borrowing regulatory sequences that cells already know how to interpret. Enhancers will also be useful for delivering potential gene therapies to patients, Wienisch says. For many years, the Feng lab has been studying how a missing copy of a gene called Shank3 impairs neurons’ ability to communicate, leading to autism and intellectual disability. Now, they are investigating whether they can overcome these deficits by delivering a functional copy of Shank3 to the brain cells that need it. Widespread activation of the therapeutic gene might do more harm than good, but incorporating the right enhancer could ensure it is delivered to the appropriate cells at the right dose, Wienisch says.
Enhancers will also be useful for delivering potential gene therapies to patients, Wienisch says. For many years, the Feng lab has been studying how a missing copy of a gene called Shank3 impairs neurons’ ability to communicate, leading to autism and intellectual disability. Now, they are investigating whether they can overcome these deficits by delivering a functional copy of Shank3 to the brain cells that need it. Widespread activation of the therapeutic gene might do more harm than good, but incorporating the right enhancer could ensure it is delivered to the appropriate cells at the right dose, Wienisch says.