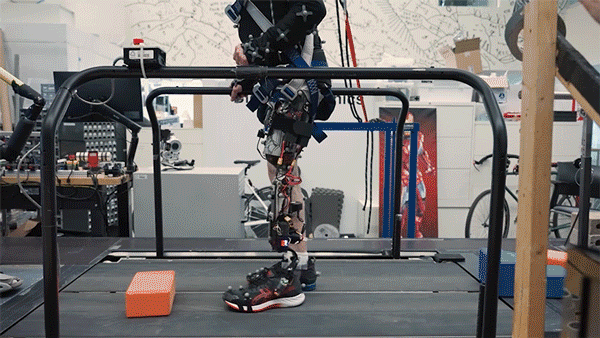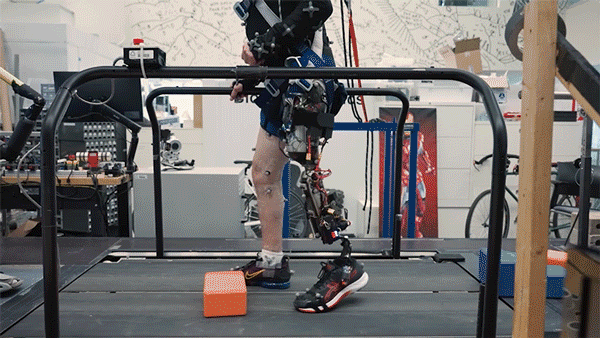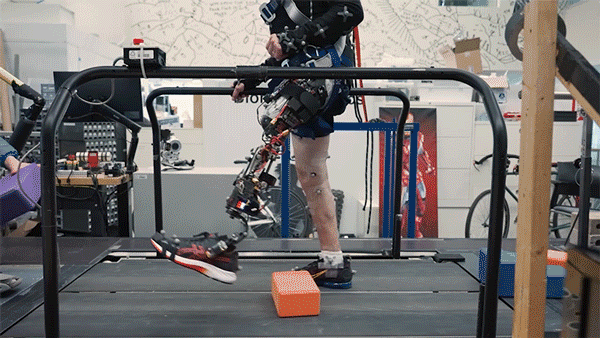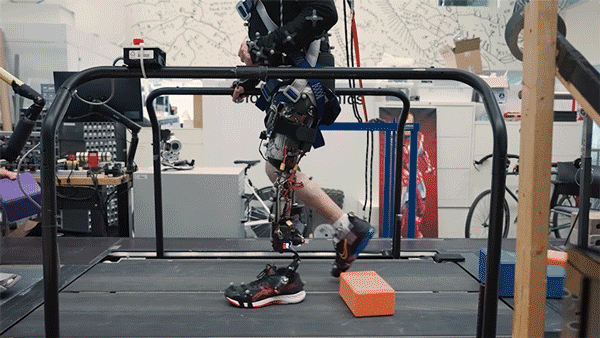
One in every eight people—970 million globally—live with mental illness, according to the World Health Organization, with depression and anxiety being the most common mental health conditions worldwide. Existing therapies for complex psychiatric disorders like depression, anxiety, and schizophrenia have limitations, and federal funding to address these shortcomings is growing increasingly uncertain.

Patricia and James Poitras ’63 have committed $8 million to the Poitras Center for Psychiatric Disorders Research to launch pioneering research initiatives aimed at uncovering the brain basis of major mental illness and accelerating the development of novel treatments.
“Federal funding rarely supports the kind of bold, early-stage research that has the potential to transform our understanding of psychiatric illness. Pat and I want to help fill that gap—giving researchers the freedom to follow their most promising leads, even when the path forward isn’t guaranteed,” says James Poitras, who is chair of the McGovern Institute Board.
Their latest gift builds upon their legacy of philanthropic support for psychiatric disorders research at MIT, which now exceeds $46 million.
“With deep gratitude for Jim and Pat’s visionary support, we are eager to launch a bold set of studies aimed at unraveling the neural and cognitive underpinnings of major mental illnesses,” says Robert Desimone, director of the McGovern Institute, home to the Poitras Center. “Together, these projects represent a powerful step toward transforming how we understand and treat mental illness.”
A legacy of support
Soon after joining the McGovern Institute Leadership Board in 2006, the Poitrases made a $20 million commitment to establish the Poitras Center for Psychiatric Disorders Research at MIT. The center’s goal, to improve human health by addressing the root causes of complex psychiatric disorders, is deeply personal to them both.
“We had decided many years ago that our philanthropic efforts would be directed towards psychiatric research. We could not have imagined then that this perfect synergy between research at MIT’s McGovern Institute and our own philanthropic goals would develop,” recalls Patricia.
The center supports research at the McGovern Institute and collaborative projects with institutions such as the Broad Institute, McLean Hospital, Mass General Brigham and other clinical research centers. Since its establishment in 2007, the center has enabled advances in psychiatric research including the development of a machine learning “risk calculator” for bipolar disorder, the use of brain imaging to predict treatment outcomes for anxiety, and studies demonstrating that mindfulness can improve mental health in adolescents.

For the past decade, the Poitrases have also fueled breakthroughs in McGovern Investigator Feng Zhang’s lab, backing the invention of powerful CRISPR systems and other molecular tools that are transforming biology and medicine. Their support has enabled the Zhang team to engineer new delivery vehicles for gene therapy, including vehicles capable of carrying genetic payloads that were once out of reach. The lab has also advanced innovative RNA-guided gene engineering tools such as NovaIscB, published in Nature Biotechnology in May 2025. These revolutionary genome editing and delivery technologies hold promise for the next generation of therapies needed for serious psychiatric illness.
In addition to fueling research in the center, the Poitras family has gifted two endowed professorships—the James and Patricia Poitras Professor of Neuroscience at MIT, currently held by Feng Zhang, and the James W. (1963) and Patricia T. Poitras Professor of Brain and Cognitive Sciences at MIT, held by Guoping Feng—and an annual postdoctoral fellowship at the McGovern Institute.
New initiatives at the Poitras Center
The Poitras family’s latest commitment to the Poitras Center will launch an ambitious set of new projects that bring together neuroscientists, clinicians, and computational experts to probe underpinnings of complex psychiatric disorders including schizophrenia, anxiety, and depression. These efforts reflect the center’s core mission: to speed scientific discovery and therapeutic innovation in the field of psychiatric brain disorders research.
McGovern cognitive neuroscientists Evelina Fedorenko PhD ‘07 and Nancy Kanwisher ’80, PhD ’86, the Walter A. Rosenblith Professor of Cognitive Neuroscience—in collaboration with psychiatrist Ann Shinn of McLean Hospital—will explore how altered inner speech and reasoning contribute to the symptoms of schizophrenia. They will collect functional MRI data from individuals diagnosed with schizophrenia and matched controls as they perform reasoning tasks. The goal is to identify the brain activity patterns that underlie impaired reasoning in schizophrenia, a core cognitive disruption in the disorder.
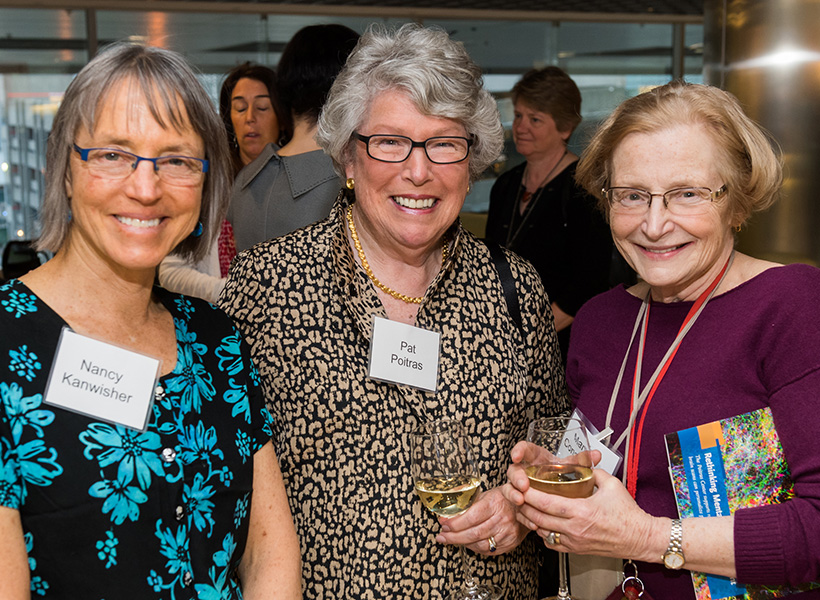
A complementary line of investigation will focus on the role of inner speech—the “voice in our head” that shapes thought and self-awareness. The team will conduct a large-scale online behavioral study of neurotypical individuals to analyze how inner speech characteristics correlate with schizophrenia-spectrum traits. This will be followed by neuroimaging work comparing brain architecture among individuals with strong or weak inner voices and people with schizophrenia, with the aim of discovering neural markers linked to self-talk and disrupted cognition.
A different project led by McGovern neuroscientist Mark Harnett and 2024–2026 Poitras Center Postdoctoral Fellow Cynthia Rais focuses on how ketamine—an increasingly used antidepressant—alters brain circuits to produce rapid and sustained improvements in mood. Despite its clinical success, ketamine’s mechanisms of action remain poorly understood. The Harnett lab is using sophisticated tools to track how ketamine affects synaptic communication and large-scale brain network dynamics, particularly in models of treatment-resistant depression. By mapping these changes at both the cellular and systems levels, the team hopes to reveal how ketamine lifts mood so quickly—and inform the development of safer, longer-lasting antidepressants.
Guoping Feng is leveraging a new animal model of depression to uncover the brain circuits that drive major depressive disorder. The new animal model provides a powerful system for studying the intricacies of mood regulation. Feng’s team is using state-of-the-art molecular tools to identify the specific genes and cell types involved in this circuit, with the goal of developing targeted treatments that can fine-tune these emotional pathways.
“This is one of the most promising models we have for understanding depression at a mechanistic level,” says Feng, who is also associate director of the McGovern Institute. “It gives us a clear target for future therapies.”
Another novel approach to treating mood disorders comes from the lab of James DiCarlo, the Peter de Florez Professor of Neuroscience at MIT, who is exploring the brain’s visual-emotional interface as a therapeutic tool for anxiety. The amygdala, a key emotional center in the brain, is heavily influenced by visual input. DiCarlo’s lab is using advanced computational models to design visual scenes that may subtly shift emotional processing in the brain—essentially using sight to regulate mood. Unlike traditional therapies, this strategy could offer a noninvasive, drug-free option for individuals suffering from anxiety.
Together, these projects exemplify the kind of interdisciplinary, high-impact research that the Poitras Center was established to support.
“Mental illness affects not just individuals, but entire families who often struggle in silence and uncertainty,” adds Patricia. “Our hope is that Poitras Center scientists will continue to make important advancements and spark novel treatments for complex mental health disorders and most of all, give families living with these conditions a renewed sense of hope for the future.”